Interested in our services
Contact our experts to provide further information



Post-translational modifications (PTMs) are essential for regulating protein activity, and pathogens use various mechanisms to interfere with host PTMs, affecting the key functions of infection. Despite the limited omics studies on macro protein PTMs, exploring microbial macro-modified proteomics can help understand the relationship between the microbiota and the host response system, decode the complex microbiome protein expression profile and modification spectrum, and establish disease models and discover drug targets. Moreover, macro-modified proteomics can also provide insights into the type and function of microorganisms in extreme environments, benefiting the research and application of environmental science.
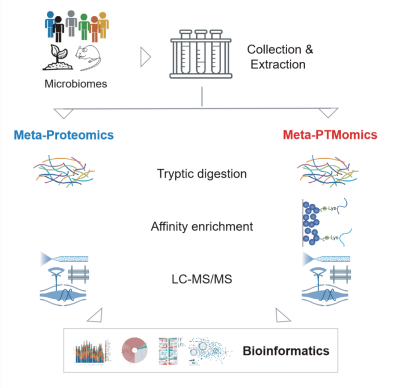
PTM-Bio has pioneered the use of mass spectrometry in macro-modified proteomics, enabling the comprehensive analysis of over ten post-translational modifications (PTMs), including acetylation and 2-hydroxyisobutyrylation. This approach allows for a systematic and in-depth investigation of the complete landscape of PTMs in microbiome proteins, thus facilitating the deciphering of complex interaction mechanisms among the microbiota, the host, and the environment.
The protein sample undergoes enzymatic digestion to generate a mixture of peptides, which is then fractionated by liquid chromatography to decrease sample complexity. Next, modified peptides are enriched using high-quality modified antibodies and biological materials, and the resulting mixture is analyzed and quantified by liquid chromatography-tandem mass spectrometry.
Inflammatory bowel disease (IBD), comprising Crohn's disease and ulcerative colitis, is a chronic and multifactorial disease that involves clinical, immune, molecular, genetic, and microbial factors, yet its etiology and pathogenesis remain incompletely understood. The gut microbiome has been widely implicated in the development of IBD, and emerging evidence suggests that microbiota transplantation represents a promising therapeutic strategy for intervention in IBD treatment.
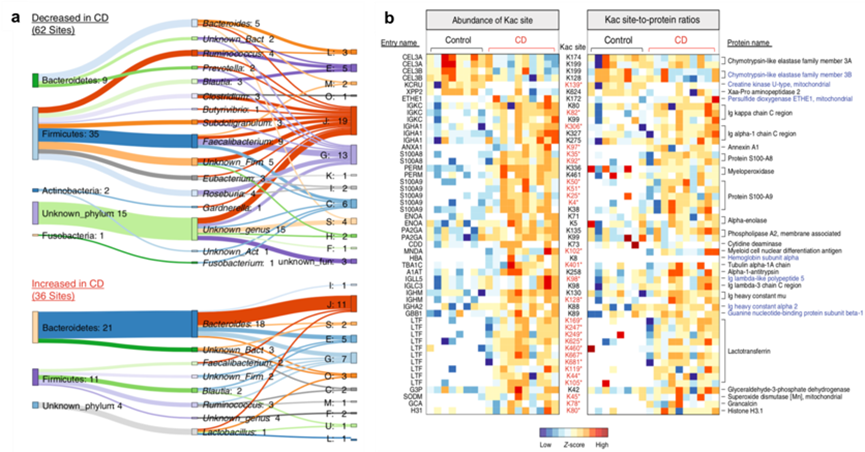
By employing a combination of mate-proteomics and mate-modified proteomics multi-omics techniques, researchers analyzed the gut microbiome of patients with Crohn's disease and uncovered alterations in acetylation modifications under disease conditions. The study revealed that acetylation modifications are prevalent in crucial metabolic pathways of the human gut microbiome, particularly in the short-chain fatty acid biosynthesis pathway in the Pachychophytes phylum. These findings highlight the pivotal role of post-translational modifications (PTMs) in functional microbiome studies.
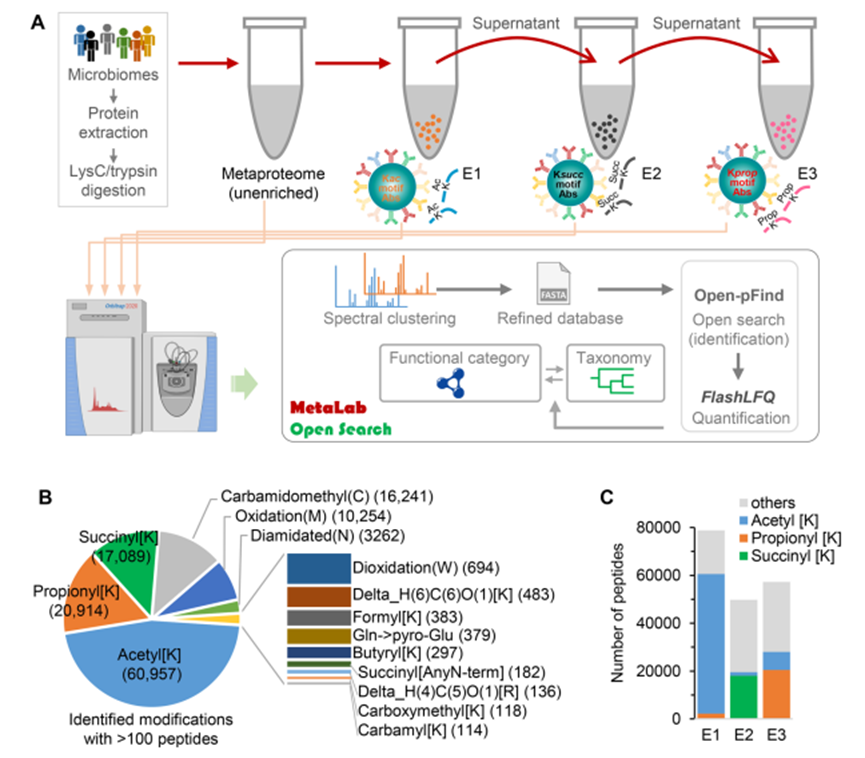
Lysine acylation modifications constitute a crucial group of post-translational modifications that are present in both eukaryotic and prokaryotic organisms, and regulate diverse cellular functions. However, our comprehension of lysine acylation modifications in the microbiome is limited by the inadequacy of effective analytical and bioinformatics tools for studying complex microbial communities.
By utilizing protein macro-modification omics techniques, the researchers conducted a comprehensive analysis of multiple lysine acylation modifications in human gut microbiome samples, identifying over 60,000 acetylated peptides, nearly 20,000 propionylated peptides, and succinylated peptides. The study revealed that lysine acylation modifications are widespread in the human gut microbiota, and can cross-talk with each other, especially in glycolytic metabolic pathways and the production of short-chain fatty acids. Moreover, most of the lysine acylated peptides originate from Phytobacterium and Bacteroides in the microbiota, and different levels of lysine acylation modifications are specifically distributed in microbial species with distinct metabolic capabilities. Macro-modified proteomics technology has significant value in the investigation of microbial species distribution and function.
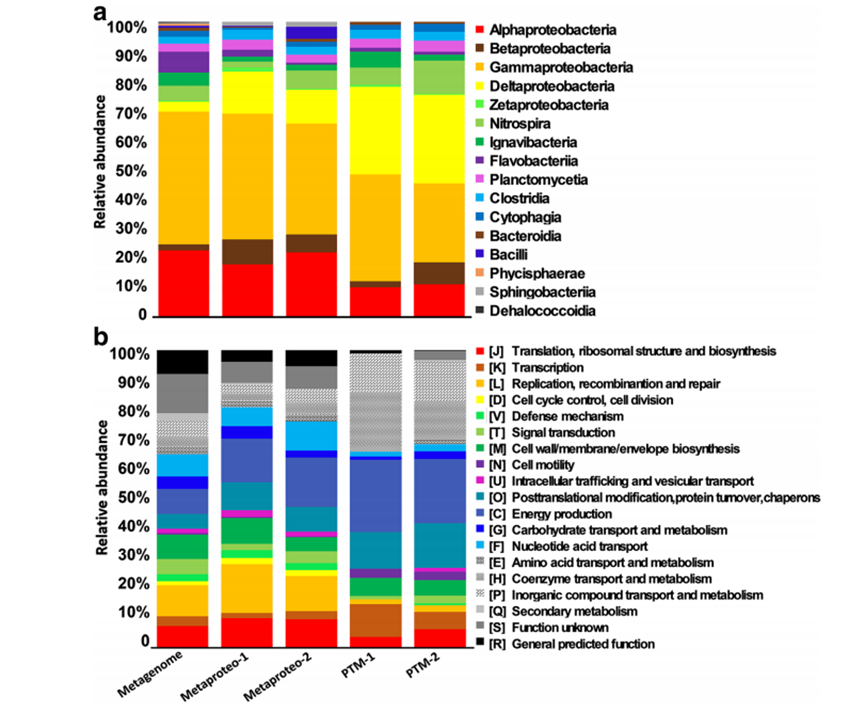
Post-translational modifications (PTMs) of proteins display significant differences between orthologous proteins from different microbial flora and are unevenly distributed across microbial taxonomy and functional categories. Through a combination of metagenomics and meta-PTMs, researchers have gained valuable insights into PTMs, including acetylation, deaminization, hydroxylation, methylation, nitrosylation, and phosphorylation in hydrothermal vent microbial communities. The study revealed that different types of modifications are unevenly distributed across microbial and functional classifications, highlighting the association between modification types and specific functions. These findings demonstrate the highly diverse and distinct PTMs of microorganisms inhabiting the deep-sea hydrothermal vents, which are critical for adaptation to extreme environmental conditions. As an essential research approach for investigating bacterial adaptation to the environment, meta-omics has significant implications in unraveling the PTM landscape in deep-sea microorganisms.
Interested in our services
Contact our experts to provide further information